[1] 4Introduction to R for Medical Students
A Practical Primer
11/02/23
Objectives
Get R/RStudio on your computer
Be able to take input data and generate output results using R
Be able to load libraries and import data
Know what a vector and dataframe are
Understand what a function is and how common functions work
Learn data wrangling basics
Getting R and RStudio
Download RStudio here
RStudio is an Integrated Development Environment (IDE), which provides a visual and interactive interface to make coding in R easier
R is the language maintained by volunteers whereas RStudio is a product maintained by a company called Posit


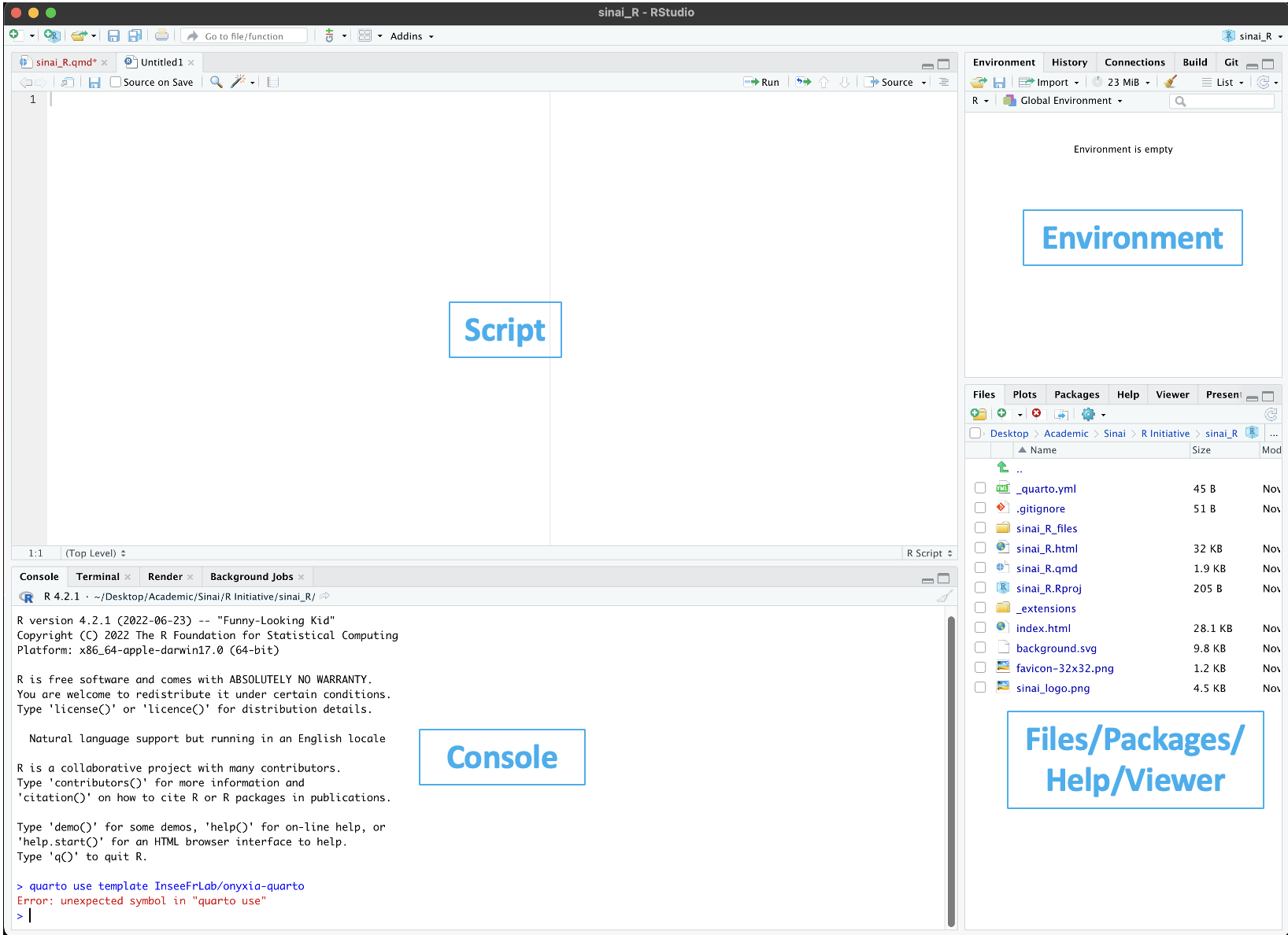
RStudio
R Packages
Base R comes installed, but power of R is being open source
install.packages("package_name")installs a package for the first time (only need to do once)The quotations “” are required
For this session, run
install.packages("tidyverse")in your console
library(package_name)loads a package into your current working session
High Yield Basic Concepts
Assignment: assign a value to an object name (e.g.
x <- 10)- Note: your name shouldn’t have spaces. Instead use snake_case or camelCase or dot.case
Functions: your “action verbs”, which take in input argument and return output (e.g.
mean())Help:
?function_name
A Little More About Functions
This is how to write your own function:
name of your function
syntax:
function() {}arguments: what you pass in as inputs
Vectors
Basic data structure in R
Function
c()combines its arguments into a vectorIndexing
[]retrieves elements of a vector by position (or by name for a named vector)Vectors can consist of numbers, characters, dates, but you cannot mix data types (e.g. numbers and characters)
structures_profs <- c("Ki Mak", "Jeffrey Laitman", "Dani Curcio")
Useful Vector Functions
length(): number of elements in vectormean(): mean of elements in vector
Be careful if you have
NAvalues (which is fairly common in most datasets)
Data Frames
Tidy data principles
Each row is an observation
Each column is a variable
Each cell contains one value
How do data frames relate to vectors?
- Imagine a data frame as a bunch of vertical vectors next to each other
data.framecreates a data frame (also look attibble)
Importing Data as Data Frames
Many formats of data
Common formats include .csv (comma), .tsv (tab), and .txt (space/tab)
Read with
readrpackage:readr::read_csv(),readr::read_tsv()orreadr::read_delim()::means namespace, which tells R in which package to look for the function
Software-specific formats include:
Excel (.xls, .xlsx)
- Read with
readxlpackage:readxl::read_excel()
- Read with
Stata (.dta)
- Read with
havenpackage:haven::read_dta()
- Read with
Viewing your Data
Either click the object in the Environment panel
![]()
Or use the
View()function (it’s cleaner to type this into your console)Use
str()to understand data types (numeric, character, date, etc.) in your dataUse
names()to view row names andcolnames()to view column names of a dataframe
Accessing your Data
How do you select specific columns?
Either use
$operator or[[ ]]operatorSepal.Length Sepal.Width Petal.Length Petal.Width Species 1 5.1 3.5 1.4 0.2 setosa 2 4.9 3.0 1.4 0.2 setosa 3 4.7 3.2 1.3 0.2 setosa 4 4.6 3.1 1.5 0.2 setosa 5 5.0 3.6 1.4 0.2 setosa 6 5.4 3.9 1.7 0.4 setosa[1] 5.1 4.9 4.7 4.6 5.0 5.4[1] 5.1 4.9 4.7 4.6 5.0 5.4
Data Wrangling Verbs You Should Know
Disclaimer: A lot of functions will be introduced in the next couple of slides, so please bear with me. We will practice these afterwards and you can always refer to the cheatsheet referenced in the last slide.
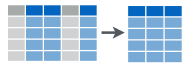
select(): select variables you want to keep![]()
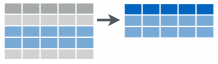
filter(): select rows you want to keep based on condition(s)![]()
![]()
mutate(): create or modify variables![]()
More Data Wrangling Verbs You Should Know
summarize(): compute summary statistics into a single row![]()
count(): tabulate counts for each level of variablegroup_by(): useful in conjuction with `summarize()` and `count()`.
Even More Data Wrangling Verbs You Should Know
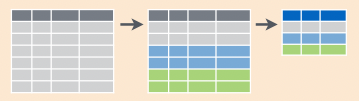
pivot_longer(): useful in conjuction with `summarize()` and `count()`![]()
separate(): useful in conjuction with `summarize()` and `count()`![]()
%>%: pipe function
How Do I Use These?
Find the average sepal length (excluding sepals greater than 7cm) for each species of iris using the iris dataset.
How Do I Use These?
Find the average sepal length (excluding sepals greater than 7cm) for each species of iris using the iris dataset.
How Do I Use These?
Find the average sepal length (excluding sepals greater than 7cm) for each species of iris using the iris dataset.
How Do I Use These?
Find the average sepal length (excluding sepals greater than 7cm) for each species of iris using the iris dataset.
How Do I Use These?
Find the average sepal length (excluding sepals greater than 7cm) for each species of iris using the iris dataset.
How Do I Use These?
Find the average sepal length (excluding sepals greater than 7cm) for each species of iris using the iris dataset.
Why %>%
Increases human readability
Makes code easier to debug
Without
%>%… yikesNote: As of R 4.0+, R comes with a built-in pipe operator
|>
Joining Dataframes
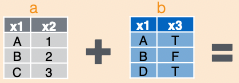
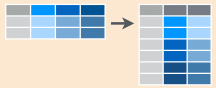
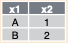
left_join(a, b, by = "x1")![]()
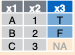
inner_join(a, b, by = "x1")![]()
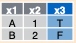
semi_join(a, b, by = "x1")![]()
- NB: You can join by multiple keys
Saving your Data
Saves a file to your computer
Use
readr::write_*()functions- The * just means a placeholder for suffixes
- Ex:
write_csv(df, "df.csv")
If you’re working with big data consider looking into the
feather,arrow, ordata.tablepackages. Base R’ssaveRDS()is also pretty serviceable.
Crap, my code’s not working
Google and StackOverflow are your best friends
- Add “R” to your search queries
Checklist for a good “reprex” (reproducible example) if you ask a question online
Provide your packages/working environment by copying/pasting output of
sessionInfo()Include your data by using
dput()and copying/pasting outputMake sure your code is as simple as possible. Only include the minimum required lines.
Ensure you’ve made a reproducible example by starting a fresh R session, pasting your script in, and running it
ChatGPT may be helpful
Acknowledgements
- Andrew Min for all his feedback on improving this presentation
Useful Highest-Yield Resources
Messaging Lathan :)
Intro to R for Data Science 2nd Edition
New pipe
|>New
dplyrsyntax